Pyrazolopyridines as potent PDE4B inhibitors: 5-heterocycle SAR.
Mitchell, C.J., Ballantine, S.P., Coe, D.M., Cook, C.M., Delves, C.J., Dowle, M.D., Edlin, C.D., Hamblin, J.N., Holman, S., Johnson, M.R., Jones, P.S., Keeling, S.E., Kranz, M., Lindvall, M., Lucas, F.S., Neu, M., Solanke, Y.E., Somers, D.O., Trivedi, N.A., Wiseman, J.O.(2010) Bioorg Med Chem Lett 20: 5803-5806
- PubMed: 20732811
- DOI: https://doi.org/10.1016/j.bmcl.2010.07.136
- Primary Citation of Related Structures:
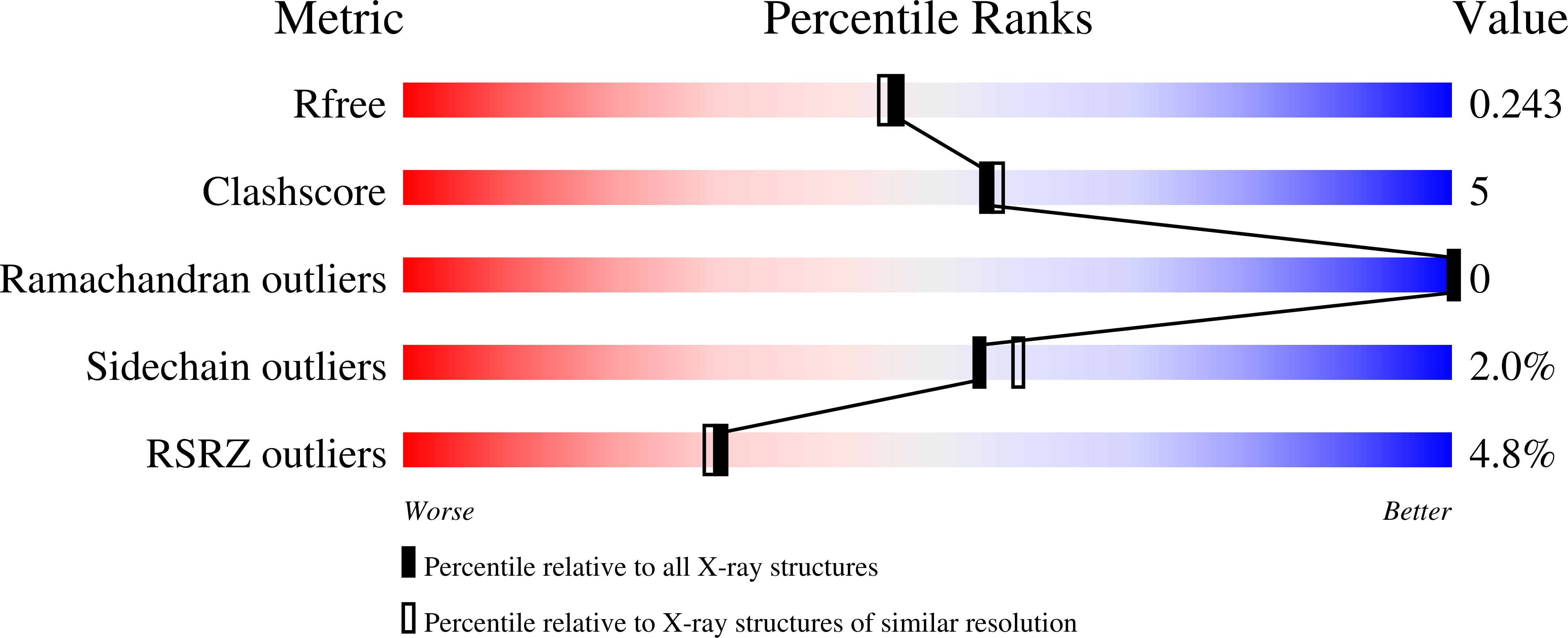
3O56, 3O57 - PubMed Abstract:
Following the discovery of 4-(substituted amino)-1-alkyl-pyrazolo[3,4-b]pyridine-5-carboxamides as potent and selective phosphodiesterase 4B inhibitors, [Hamblin, J. N.; Angell, T.; Ballentine, S., et al. Bioorg. Med. Chem. Lett.2008, 18, 4237] the SAR of the 5-position was investigated further. A range of substituted heterocycles showed good potencies against PDE4. Optimisation using X-ray crystallography and computational modelling led to the discovery of 16, with sub-nM inhibition of LPS-induced TNF-α production from isolated human peripheral blood mononuclear cells.
Organizational Affiliation:
GlaxoSmithKline, Medicines Research Centre, Gunnels Wood Road, Stevenage, Hertfordshire SG1 2NY, United Kingdom. charlotte.j.mitchell@gsk.com